Posts
How different are Cats vs. Cells in Histopathology?
/in Science News/by Andreas HolzingerAn awesome question stated in an article by Michael BEREKET and Thao NGUYEN (Febuary 7, 2018) brings it straight to the point: Deep learning has revolutionized the field of computer vision. So why are pathologists still spending their time looking at cells through microscopes?
The most famous machine learning experiments have been done with recognizing cats (see the video by Peter Norvig) – and the question is relevant, how different are these cats from the cells in histopathology?
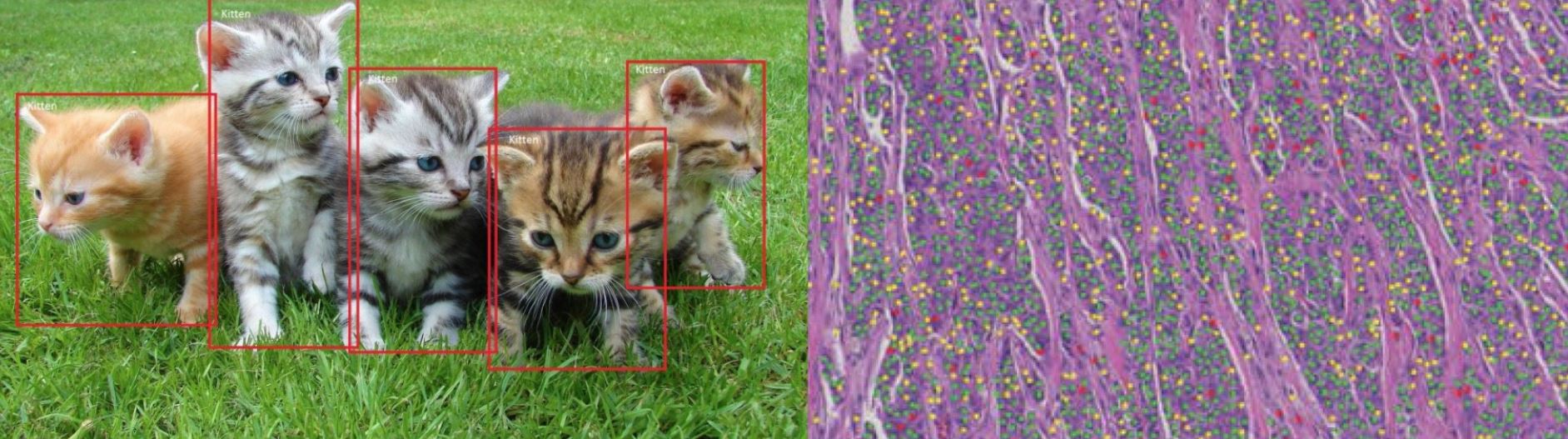
Machine Learning, and in particular deep learning, has reached a human-level in certain tasks, particularly in image classification. Interestingly, in the field of pathology these methods are not so ubiqutiously used currently. A valid question indeed is: Why do human pathologists spend so much time with visual inspection? Of course we restrict this debate on routine tasks!
This excellent article is worthwhile giving a read:
Stanford AI for healthcare: How different are cats from cells
Source of the animated gif above:
https://giphy.com/gifs/microscope-fluorescence-mitosis-2G5llPaffwvio
“Machine Learning for Health Informatics” Lecture Notes in Artificial Intelligence 9605 > 40,626 downloads 2017
/in HCI-KDD Events, Science News/by Andreas HolzingerSince its online publication on December 10, 2016 the Volume edited by Andreas Holzinger “Machine Learning for Health Informatics” Springer Lecture Notes in Artificial Intelligence LNAI Volume 9605, has been downloaded 54,960 times as of today (May, 11, 2018, 20:00 CEST) and 44,988 with status as of April 2018 according to the official Springer Bookmetrix book performance report – a record; and alone in the year 2017 40,626 downloads, which is 10 times higher than a typical volume of the series of Lecture Notes in Artificial Intelligence by Springer/Nature. A cordial thank you for my international colleagues for this huge acceptance!
https://www.springer.com/gp/book/9783319504773
A popular passage from the book:
https://books.google.com/talktobooks/query?q=What%E2%80%99s%20the%20difference%20between%20Machine%20Learning%20and%20deep%20learning%3F
Update on 15th September 2018: 63k downloads
AI will change Radiology – NOT replace Radiologists
/in Science News/by Andreas Holzinger After the rather shocking statement of Geoffrey HINTON during the Machine Learning and Market for Intelligence Conference in Toronto, where he recommended that hospitals should stop training radiologists, because deep learning will replace them (watch video below), on March, 27, 2018 Thomas H. DAVENPORT and Keith J. DREYER published a really nice article on “AI will change radiology, but it won’t replace radiologists” (see [1]) – which supports our human-in-the-loop approach: for sure, AI/machine learning (difference here) will change workflows, but we envision that the expert will be augmented by new technologies, i.e. routine (boring) tasks will be replaced by automatic algorithms, but this will free up expert time to spent on challenging (cool) tasks and more research – and there are plenty of problems where we need human intelligence!
After the rather shocking statement of Geoffrey HINTON during the Machine Learning and Market for Intelligence Conference in Toronto, where he recommended that hospitals should stop training radiologists, because deep learning will replace them (watch video below), on March, 27, 2018 Thomas H. DAVENPORT and Keith J. DREYER published a really nice article on “AI will change radiology, but it won’t replace radiologists” (see [1]) – which supports our human-in-the-loop approach: for sure, AI/machine learning (difference here) will change workflows, but we envision that the expert will be augmented by new technologies, i.e. routine (boring) tasks will be replaced by automatic algorithms, but this will free up expert time to spent on challenging (cool) tasks and more research – and there are plenty of problems where we need human intelligence!
[1] https://hbr.org/2018/03/ai-will-change-radiology-but-it-wont-replace-radiologists
Call for Papers: Open Data for Discovery Science (due to July, 31, 2017)
/in Calls for Papers, HCI-KDD Events/by Andreas HolzingerThe Journal BMC Medical Informatics and Decision Making (SCI IF (2015): 2,042)
invites to submit to a new thematic series on open data for discovery science
https://bmcmedinformdecismak.biomedcentral.com/articles/collections/odds
Note: Excellent submissions to the IFIP Cross Domain Conference on Machine Learning and Knowledge Discovery (CD-MAKE), (Submission due to May, 15, 2017) relevant to the topics described below, will be invited to expand their work into this thematic series:
The use of open data for discovery science has gained much attention recently as its full potential is unfolding and being explored in projects spanning all areas of healthcare research. A plethora of data sets are now available thanks to drives to make data universally accessible and usable for discovery science. However, with these advances come inherent challenges with the processing and management of ever expanding data sources. The computational and informatics tools and methods currently used in most investigational settings are often labor intensive and rely upon technologies that have not been designed to scale and support reasoning across multi-dimensional data resources. In addition, there are many challenges associated with the storage and responsible use of open data, particularly medical data, such as privacy, data protection, safety, information security and fair use of the data. There are therefore significant demands from the research community for the development of data management and analytic tools supporting heterogeneous analytic workflows and open data sources. Effective anonymisation tools are also of paramount importance to protect data security whilst preserving the usability of the data.
The purpose of this thematic series is to bring together articles reporting advances in the use of open data including the following:
- The development of tools and methods targeting the reproducible and rigorous use of open data for discovery science, including but not limited to: syntactic and semantic standards, platforms for data sharing and discovery, and computational workflow orchestration technologies that enable the creation of data analytics, machine learning and knowledge extraction pipelines.
- Practical approaches for the automated and/or semi-automated harmonization, integration, analysis, and presentation of data products to enable hypothesis discovery or testing.
- Theoretical and practical approaches for solutions to make use of interactive machine learning to put a human-in-the-loop, answering questions including: could human intelligence lead to general heuristics that we can use to improve heuristics?
- Frameworks for the application of open data in hypothesis generation and testing in projects spanning translational, clinical, and population health research.
- Applied studies that demonstrate the value of using open data either as a primary or as an enriching source of information for the purposes of hypothesis generation/testing or for data-driven decision making in the research, clinical, and/or population health environments.
- Privacy preserving machine learning and knowledge extraction algorithms that can enable the sharing of previously “privileged” data types as open data.
- Evaluation and benchmarking methodologies, methods and tools that can be used to demonstrate the impact of results generated through the primary or secondary use of open data.
- Socio-cultural, usability, acceptance, ethical and policy issues and frameworks relevant to the sharing, use, and dissemination of information and knowledge derived from the analysis of open data.
Submission is open to everyone, and all submitted manuscripts will be peer-reviewed through the standard BMC Medical Informatics and Decision Making review process. Manuscripts should be formatted according to the submission guidelines and submitted via the online submission system. Please indicate clearly in the covering letter that the manuscript is to be considered for the ‘Open data for discovery science’ collection. The deadline for submissions will be 31 July 2017.
For further information, please email the editors of the thematic series:
Andreas HOLZINGER a.holzinger@human-centered.ai,
Philip PAYNE prpayne@wustl.edu ,or the BMC in-house editor
Emma COOKSON at emma.cookson@biomedcentral.com
Link to the IFIP Cross-Domain Conference on Machine Learning and Knowledge Extraction (CD-MAKE):
https://cd-make.net
Integrated interactomes and pathways in precision medicine by Igor Jurisica, Toronto
/in HCI-KDD Events, Lectures/by Andreas HolzingerMachine learning is the fastest growing field in computer science, and Health Informatics is amongst the greatest application challenges, providing benefits in improved medical diagnoses, disease analyses, and pharmaceutical development – towards future precision medicine.
Talk announcement: Friday, 12th May, 2017, 10:00, Seminaraum 137, Parterre, Inffeldgasse 16c
Integrated interactomes and pathways in precision medicine
by Igor Jurisica, University of Toronto and Princess Margaret Cancer Center Toronto
Abstract: Fathoming cancer and other complex disease development processes requires systematically integrating diverse types of information, including multiple high-throughput datasets and diverse annotations. This comprehensive and integrative analysis will lead to data-driven precision medicine, and in turn will help us to develop new hypotheses, and answer complex questions such as what factors cause disease; which patients are at high risk; will patients respond to a given treatment; how to rationally select a combination therapy to individual patient, etc.
Thousands of potentially important proteins remain poorly characterized. Computational biology methods, including machine learning, knowledge extraction, data mining and visualization, can help to fill this gap with accurate predictions, making disease modeling more comprehensive. Intertwining computational prediction and modeling with biological experiments will lead to more useful findings faster and more economically.
Short Bio: Igor Jurisica is Tier I Canada Research Chair in Integrative Cancer Informatics, Senior Scientist at Princess Margaret Cancer Centre, Professor at University of Toronto and Visiting Scientist at IBM CAS. He is also an Adjunct Professor at the School of Computing, Pathology and Molecular Medicine at Queen’s University, Computer Science at York University, scientist at the Institute of Neuroimmunology, Slovak Academy of Sciences and an Honorary Professor at Shanghai Jiao Tong University in China. Since 2015, he has also served as Chief Scientist at the Creative Destruction Lab, Rotman School of Management. Igor has published extensively on data mining, visualization and cancer informatics, including multiple papers in Science, Nature, Nature Medicine, Nature Methods, Journal of Clinical Oncology, and received over 9,960 citations since 2012. He has been included in Thomson Reuters 2016, 2015 & 2014 list of Highly Cited Researchers, and The World’s Most Influential Scientific Minds: 2015 & 2014 Reports.
Jurisica Lab, IBM Life Sciences Discovery Center:
Canada Tier I Research Chair: https://www.chairs-chaires.gc.ca/chairholders-titulaires/profile-eng.aspx?profileId=2347
On Nutrigenomics [1]: https://www.uhn.ca/corporate/News/Pages/Igor_Jurisica_talks_nutrigenomics.aspx
[1] Nutrigenomics tries to define the causality or relationship between specific nutrients and specific nutrient regimes (diets) on human health. The underlying idea is in personalized nutrition based on the *omics background, which may help to foster personal dietrary recommendations. Ultimately, nutrigenomics will allow effective dietary-intervention strategies to recover normal homeostasis and to prevent diet-related diseases, see: Muller, M. & Kersten, S. 2003. Nutrigenomics: goals and strategies. Nature Reviews Genetics, 4, (4), 315-322.
Call for Papers – Privacy Aware Machine Learning PAML due to April, 1, 2017
/in Calls for Papers, HCI-KDD Events/by Andreas HolzingerPrivacy Aware Machine Learning (PAML)
for Health Data Science
Special Session on September, 1, 2017, organized by Andreas HOLZINGER, Peter KIESEBERG, Edgar WEIPPL and A Min TJOA in the context of the 12th International Conference on Availability, Reliability and Security (ARES and CD-ARES), Reggio di Calabria, Italy, August 29 – September, 2, 2017
supported by the International Federation of Information Processing IFIP > TC5 and WG 8.4 and WG 8.9
https://cd-ares-conference.eu
https://www.ares-conference.eu
Keynote Talk by Neil D. LAWRENCE, University of Sheffield and Amazon
With the new European data protection and privacy regulations coming into effect with January, 1, 2018 issues having been nice to have so far are becoming a must have. Privacy aware machine learning will be one of the most important fields for the European research community and the IT business in particular. Most affected is the whole area of biology, medicine and health, partiuclarly driven by the fact that health sciences are becoming a more and more data intensive science.
This special session will bring together scientists with diverse background, interested in both the underlying theoretical principles as well as the application of such methods for practical use in the biomedical, life sciences and health care domain. The cross-domain integration and appraisal of different fields will provide an atmosphere to foster different perspectives and opinions; it will offer a platform for novel crazy ideas and a fresh look on the methodologies to put these ideas into business.
All paper will be peer-reviewed by three members of the international PAML-commitee. Paper acceptance rate of the last session was 35 %. Accepted papers will be published in a Springer Lecture Notes in Computer Science (LNCS) Volume and excellent contributions will be invited to be extented in a special issue of a journal (planned Springer MACH and/or BMC MIDM).
Research topics covered by this special session include but are not limited to the following topics:
– Production of Open Data Sets
– Synthetic data sets for learning algorithm testing
– Privacy preserving machine learning, data mining and knowledge discovery
– Data leak detection
– Data citation
– Differential privacy
– Anonymization and pseudonymization
– Securing expert-in-the-loop machine learning systems
– Evaluation and benchmarking
This picture was taken by our local host, Francesco Buccafurri on January, 3, 2017: from the conference venue you have a direct view to the Aetna volcano:
3,2 Trillion USD on health per year
/in Science News/by Andreas HolzingerThe U.S. spends more on health care than any other country
Dieleman et al. (2016) just (Dec, 27, 2016) published a paper [1] which discusses data from the National Health Expenditure Accounts to estimate US spending on personal health care and public health, according to condition, age and sex group, and type of care. This paper was mentioned in the Washington Post by Carolyn Y. Johnson on December 27 at 11:00 AM
Here a link to the original paper:
[1] Dieleman JL, Baral R, Birger M, Bui AL, Bulchis A, Chapin A, Hamavid H, Horst C, Johnson EK, Joseph J, Lavado R, Lomsadze L, Reynolds A, Squires E, Campbell M, DeCenso B, Dicker D, Flaxman AD, Gabert R, Highfill T, Naghavi M, Nightingale N, Templin T, Tobias MI, Vos T, Murray CJL. US Spending on Personal Health Care and Public Health, 1996-2013. JAMA. 2016;316(24):2627-2646. doi:10.1001/jama.2016.16885
Here the article (shortened) from the Washington Post:
American health-care spending, measured in trillions of dollars, boggles the mind. Last year, we spent $3.2 trillion on health care a number so large that it can be difficult to grasp its scale.
A new study published in the Journal of the American Medical Association reveals what patients and their insurers are spending that money on, breaking it down by 155 diseases, patient age and category — such as pharmaceuticals or hospitalizations. Among its findings:
- Chronic — and often preventable — diseases are a huge driver of personal health spending. The three most expensive diseases in 2013: diabetes ($101 billion), the most common form of heart disease ($88 billion) and back and neck pain ($88 billion).
- Yearly spending increases aren’t uniform: Over a nearly two-decade period, diabetes and low back and neck pain grew at more than 6 percent per year — much faster than overall spending. Meanwhile, heart disease spending grew at 0.2 percent.
- Medical spending increases with age — with the exception of newborns. About 38 percent of personal health spending in 2013 was for people over age 65. Annual spending for girls between 1 and 4 years old averaged $2,000 per person; older women 70 to 74 years old averaged $16,000.
Here the link to the original article:
https://www.washingtonpost.com/news/wonk/wp/2016/12/27/the-u-s-spends-more-on-health-care-than-any-other-country-heres-what-were-buying/?tid=pm_business_pop&utm_term=.71fc517cdc11
LNAI 9605 Machine Learning for Health Informatics available
/in Recent Publications, Science News/by Andreas Holzinger14.12.2016 LNAI 9605 just appeared
Machine Learning for Health Informatics Lecture Notes in Artificial Intelligence LNAI 9605
Holzinger, Andreas (ed.) 2016. Machine Learning for Health Informatics: State-of-the-Art and Future Challenges. Cham: Springer International Publishing, doi:10.1007/978-3-319-50478-0
Machine learning (ML) is the fastest growing field in computer science, and Health Informatics (HI) is amongst the greatest application challenges, providing future benefits in improved medical diagnoses, disease analyses, and pharmaceutical development. However, successful ML for HI needs a concerted effort, fostering integrative research between experts ranging from diverse disciplines from data science to visualization.
Tackling complex challenges needs both disciplinary excellence and cross-disciplinary networking without any boundaries. Following the HCI-KDD approach, in combining the best of two worlds, it is aimed to support human intelligence with machine intelligence.
This state-of-the-art survey is an output of the international HCI-KDD expert network and features 22 carefully selected and peer-reviewed chapters on hot topics in machine learning for health informatics; they discuss open problems and future challenges in order to stimulate further research and international progress in this field.














